sddsvexperiment varies process variables and measures process variables, with optional averaging.
An input file of namelist commands gives the specific instructions.
The results are recorded in one or more SDDS files. This command differs from
sddsexperiment in that the measurement process variables are specified in two lists. The first
list gives PV ``rootnames''. The second list gives suffixes to apply to each of
the rootnames.
sddsvexperiment S1A:H1.vexp S1A:H1.sdds
where the contents of the file S1A:H1.exp are
&rootname_list
filename=bpmRoot.sdds
&end
&suffix_list
filename=bpmSuffix.sdds
&end
&variable PV_name = "S1A:H1:CurrentAO",
parameter_name="S1A:H1"
! the corrector is varied in 5 steps from -1.0 to 1.0 amps.
index_number = 0, index_limit = 5,
initial_value = -1.0, final_value = 1.0,
&end
&execute
post_change_pause=4,
intermeasurement_pause=1
&end
where the line starting with a ``!'' is a comment.
The contents of the file bpmRoot.sdds is
SDDS1 &description &end &column name = "Rootname", type = "string", &end &data mode = "ascii", &end 360 ! number of rows S1A:P1 S1A:P2 S1A:P3 S1A:P4 S1B:P5 S1B:P4 S1B:P3 S1B:P2 S1B:P1 S2A:P1 ...The contents of the file
bpmSuffix.sdds is
SDDS1 &description &end &column name=Suffix, type=string &end &column name=NumberToAverage, type=long &end &data mode=ascii, no_row_counts=1 &end :ms:x 10 :ms:y 10
usage: sddsvexperiment <inputFile> <outputFile> [-suffixFile=<filename>] [-rootnameFile=<filename>] [-echoinput] [-dryrun] [-summarize] [-verbose[=very]] [-ezcaTiming=<timeout>,<retries>] [-describeInput]
- input file:
The input file consists of namelist commands that set up and execute the experiment. The functions of the commands are described below.
variable-- specifies a process variable to vary, and the range and steps of the variation. More than one variable command may be defined, so that many process variables may vary at a time.rootname_list-- specifies rootnames from which to generate process variable to measure at each step during the experiment.suffix_list-- specifies suffixes from which to generate process variable to measure at each step during the experiment.execute-- start executing the experiment. One group of variable, measurement and execute commands may follow another in the same file for multiple experiments.erase-- deletes previous variable or measurement setups.list_control_quantities-- makes a cross-reference file for process variable names and column names of the data file.system_call-- specifies a system call (usually a script) to be executed either before a measurement or before setting a process variable.
&<command-name> <variable-type> <variable-name> = <default-value> . . . &endwhere the part
<variable-type>, which doesn't appear in an actual command, is used to illustrate the valid type of the value. The three valid types are:double-- for a double-precision type, used for most physical quantity variables,long-- for an integer type, used for flags mostly.STRING-- for a character string enclosed in double quotes.
&<command-name> [<variable-name> = <value>,] ... &endIn the namelist definition listings the square brackets denotes an optional component. Not all variables need to be defined - the defaults may be sufficient. Those that do need to be defined are noted in the detailed explanations. The only variables that don't have default values in general are string variables.
variable- function: Specifies a process variable to vary, and the range
and steps of the variation. Values of variables at each
measurement step are written to an SDDS output file. The
readback-related fields are used to confirm that the physical
device has responded to a setpoint command at every step (and
substep) within some tolerance. Readback is enabled when
readback_attemptsandreadback_toleranceare defined with non-zero positive values.When an arbitrary sequence of setpoint values is required (say a binary sequence), the values can be read in from an SDDS file specified by the
values_filefield. The fields associated for the range and steps are ignored in this case.With multiple
variablecommands, variables may be varied in a multi-dimensional grid. For example, variables may be varied independently of each other, or some groups of variables may vary together forming one axis of a multi-dimensional grid (see itemindex_number).&variable STRING PV_name = NULL STRING parameter_name = NULL STRING symbol = NULL STRING units = "unknown" double initial_value = 0 double final_value = 0 long relative_to_original = 0 long index_limit = 0 long index_number = 0 STRING function = NULL STRING values_file = NULL; STRING values_file_column = NULL; long substeps = 1 double substep_pause = 0 double range_multiplier = 1 STRING readback_name = NULL double readback_pause = 0.1 double readback_tolerance = 0 long readback_attempts = 10 long reset_to_original = 1 &end PV_name-- Required. Process variable name to vary.parameter_name-- Required. Parameter name for the variable data recorded in the output file.symbol-- Optional. Symbol field for the above column definition of the variable data.units-- Optional. Units field for the above column definition of the variable data.initial_value-- Required. The initial value of the process variable in the variation.final_value-- Required. The final value of the process variable in the variation.index_limit-- Number of steps in the variation. Measurements are taken at each step.index_number-- Required. The counter (or index) number with which the defined variation is associated. In asddsexperimentrun, counters must be defined in an increasing sequence starting from counter 0. That is, the firstvariablecommand of the file must haveindex_number= 0. The secondvariablecommand must haveindex_number= 0 or 1. In the former case, the two variables will move together with the same number of steps according their respectiveinitial_valueandfinal_value. In the latter case, the two variables will vary independently of each other with possibly different number of steps in a 2-dimensional grid.Counter number
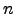 is nested within counter
is nested within counter 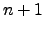 . Therefore it
might be efficient to assign devices with slower response times
to higher
. Therefore it
might be efficient to assign devices with slower response times
to higher index_numbercounter.index_limit-- Normally required. Number of steps in the variation. Measurements are taken at each step. When more than one variable is associated with the same counter, only theindex_limitof the first variable definition for that counter need to be defined. Ifindex_limitis defined invariablecommands of the sameindex_numbervalue, then the firstindex_limitremain in force.relative_to_original-- Optional. If non-zero, then the variation range is defined relative to the original process variable value (i.e. the value prior to running the program).range_multiplier-- Optional. Factor by which the range,final_value-initial_value, is multiplied. New values ofinitial_valueandfinal_valueare calculated while keeping the midpoint of the range the same.function-- Optional. A string of rpn operations used to transform the range specified byinitial_value,final_value, andindex_limit. For convenience, the original value of the process variable, and the calculated grid value for the process variable on the current step or substep are automatically pushed onto the the stack before the function is executed. The calculated values are recorded in the output file. The environment variableRPN_DEFNSis used to read a rpn definition file at the start of the execution ofsddsvexperiment.values_file-- Optional. An SDDS data file containing setpoints for the variable. This is useful is one has arbitrary setpoints values to apply. The values of the fieldsinitial_value,final_value,_substeps,range_multiplierandindex_limitare ignored.One can have other
variablenamelists with the sameindex_numberthat don't use a file for the values. The defaultindex_limitof the other variable will be set to the number of setpoint in the values file. Thus the values in the file and the values calculated for the other variable will vary together with the same number of steps.values_file_column-- Required whenvalues_fileis specified.values_file_columngives the column name of the setpoints data in filevalues_file.substeps-- Optional. If greater than one, the steps are subdivided into this number. Measurements are not made at substeps. Substeps are useful when the physical device associated with the process variable cannot reliably make steps as large as those that might be defined withinitial_value,final_value, andindex_limit.substep_pause-- Optional. Number of seconds to pause after the variable change of each substeps.readback_name-- Optional. Readback process variable name associated withPV_name. The default value forreadback_nameisPV_name.readback_tolerance-- Optional. Maximum acceptable absolute value of the difference between the process variable setpoint and its readback. A positive value is required in order to enable readbacks.readback_pause-- Optional. Number of seconds to pause after each reading of thereadback_nameprocess variable. This pause time is in addition to other pauses defined.readback_attempts-- Optional. Number of allowed readings of thereadback_nameprocess variable and readback pauses after a variable change has occured. After this number of readings, the program exits. The first readback is attempted immediately (i.e. no pause) after sending a setpoint command to thePV_name. A positive value is required in order to enable readbacks.reset_to_original-- Optional. A value of 1 means that the variable is reset to its original value when the experiment terminates normally or abnormally.
- function: Specifies a process variable to vary, and the range
and steps of the variation. Values of variables at each
measurement step are written to an SDDS output file. The
readback-related fields are used to confirm that the physical
device has responded to a setpoint command at every step (and
substep) within some tolerance. Readback is enabled when
rootname_list- function: specifies rootnames from which to generate names of process variable to measure at each step during the experiment.
&rootname_list STRING filename = NULL; &end filename-- Required. See description of rootname file below.
- function: specifies rootnames from which to generate names of process variable to measure at each step during the experiment.
suffix_list- function: specifies suffixes from which to generate names of process variable to measure at each step during the experiment.
&suffix_list STRING filename = NULL; &end filename-- Required. See description of suffix file below.
- function: specifies suffixes from which to generate names of process variable to measure at each step during the experiment.
execute- function: start executing the experiment. Some global parameters are defined here.
&execute double post_change_pause = 0 double intermeasurement_pause = 0 double rollover_pause = 0 long post_change_key_wait = 0 long allowed_timeout_errors = 1 long allowed_limit_errors = 1 double outlimit_pause = 0.1 long repeat_reading = 1 double post_reading_pause = 0.1 double ramp_pause = 0.25; long ramp_steps = 10; &end post_change_pause-- Optional. Number of seconds to pause after each change before attempting to make any measurement.intermeasurement_pause-- Optional. Number of seconds to pause between each measurement. Individual measurements for averaging are taken at this interval.rollover_pause-- Optional. Number of seconds to pause after a counter has reached its upper limit, and must rollover to zero. This allows any physical devices associated with the counter to settle after a change equal to the total range of the variation.post_change_key_wait-- Optional. If non-zero, then wait for a key press after making variable changes but before taking measurements. A prompt is given.allowed_timeout_errors-- Optional. Number of timeout errors allowed before aborting the program.allowed_limit_errors-- Optional. Number of invalid range measurement errors allowed before aborting the program. The valid range of a measurement is specified in themeasurementcommand.outlimit_pause-- Optional. Number of seconds to pause after an invalid range measurement error occured. This is to permit equipment time to recover from whatever glitch caused the out-of-limit reading.repeat_reading-- Optional. The measurements and statistical analyses are repeated this number of times for each variable settings. A page of data is written to the output file for each repitition.post_reading_pause-- Optional. Number of seconds to pause after taking a set of measurements and making a statistical analysis. If measurements are repeated then the pause is repeated after each set of measurements.ramp_steps-- Optional. Number of steps in the variables PV ramp which occurs at the start and the end of the experiment.Ramping is necessary for some devices that do not respond well to large changes to their setpoints. Ramping is done at the start of the experiments to slowly change the variable PVs from their current values to their initial values. Another ramp is done at the end to slowly bring the variable PVs from their final values back the original values. Ramping back to original values is also done when the experiment aborts for some reason.
ramp_pause-- Optional. Time interval at each step of the variables PV ramp which occurs at the start and the end of the experiment. This is not the same variable as the pause between variable changes during the experiment.
- function: start executing the experiment. Some global parameters are defined here.
erase- function: deletes previous variable or measurement setups.
&erase long variable_definitions = 1 long measurement_definitions = 1 &end variable_definitions-- Optional. If non-null, then all the variable definitions are erased.measurement_definitions-- Optional. If non-null, then all the measurement definitions are erased.
- function: deletes previous variable or measurement setups.
list_control_quantities- function: makes a cross-reference file for process variable names and column names of the data file.
&list_control_quantities STRING filename = NULL &end filename-- Required. Name of file. Columns defined areControlName,SymbolicName, andControlUnits.
- function: makes a cross-reference file for process variable names and column names of the data file.
system_call- function: specifies a system call (usually a script) to be executed repeatedly during the experiment.
&system_call STRING command = NULL long index_number = 0 long index_limit = 0 double post_command_pause = 0 double pre_command_pause = 0 long append_counter = 0 STRING counter_format = "%ld" long call_before_setting = 0 long call_before_measuring = 1 STRING counter_column_name = NULL &end command-- Required. Name of shell command or script to execute.index_number-- Required. Counter number with which the command will be associated. The command is executed when this counter is advanced or rolled over.index_number-- Optional. Number of times the command is executed for the associated counter. This field is used only when the value ofindex_numberabove defines a new counter.post_command_pause-- Optional. Number of seconds to pause after the completion of the command.pre_command_pause-- Optional. Number of seconds to pause before executing the command.append_counter-- Optional. If non-zero, the counter value is appended to the command when the system call is made.counter_format-- Optional. Format for the counter if the counter value is appended to the command.call_before_setting,call_before_measuring, -- Optional. At a counter advance or rollover the command can be executed in one of three ways:- before both variable changes and measurements:
call_before_setting=1,call_before_measuring=1 - after variable changes and before measurements:
call_before_setting=0,call_before_measuring=1 - after both variable changes and measurements:
call_before_setting=0,call_before_measuring=0
- before both variable changes and measurements:
counter_column_name-- Optional. If non-null, a column in the output file with this name is defined. The values written to this column are the number of times the command had been called minus one. This value doesn't rollover with its associated counter.
- function: specifies a system call (usually a script) to be executed repeatedly during the experiment.
- Rootname file:
SDDS file defining the rootnames of the process variables with colum:
Rootname-- Required string column to generate measurement process variables.
- Suffix file:
SDDS file defining the suffixes and measurement parameters with columns:
Suffix-- Required string columne of suffix names to be appended to theRootnamevalues in the rootname file to generate measurement process variables.ColumnNameSuffix-- Optional string column of names of suffix column appearing in output data file.NumberToAverage-- Optional long column of number of measurements to average.IncludeStDev-- Optional character column. If value is ``y'' then the standard deviation (a measure of the distribution of measurements) is calcualted and included in the output file. If ``n'' then the standard deviation is not calculated.IncludeSigma-- Optional character column. If value is ``y'' then the sigma (uncertainty on the mean value) is calcualted and included in the output file. If ``n'' then the sigma is not calculated.LowerLimitandUpperLimit-- Optional double columns. Must have both or neither. Defines a range of validity for the individual measurements. If the number of invalid measurements (reset to 0 at each measurement step) equals or exceeds the value ofallowed_limit_errors(default of 1) in commandexecute, then the program aborts. The average values written to the output file excludes measurements outside this range.
- Output file:
The output file contains one data page for each variable step. The names of the defined columns are those string data of the
SuffixorColumnNameSuffixcolumns from the suffix file. A column is created for each standard deviation or sigma calculation requested for a measurement. The standard deviation columns are namedStDev<columnName>, and the sigma columns are namedStDev<columnName>, where<columnName>is replaced by an actual column name.Some additional columns are defined:
Rootname-- String column for the rootname of the PVs.Index-- Long columns for the index of the row.- Time -- Double parameter of time since start of epoch. This time data can be used by
the plotting program
sddsplotto make the best coice of time unit conversions for time axis labeling. - ElapsedTime -- Double parameter of elapsed time of readback since the start of the experiment.
The variable values appear as parameters in each data page.
Many time parameters are defined:
Step-- Long parameter for the step number.- Time -- Double parameter of time since start of epoch. This time data can be used by
the plotting program
sddsplotto make the best coice of time unit conversions for time axis labeling. - ElapsedTime -- Double parameter of elapsed time of readback since the start of the experiment.
- TimeOfDay -- Double parameter of system time in units of hours. The time does not wrap around at 24 hours.
- TimeStamp -- String parameter of time stamp for file.
-suffixFile=<filename>-- SDDS file defining the suffixes and measurement parameters. If not specified on the command line, then the namelist commandsuffix_listis required in the input file.-rootnameFile=<filename>-- SDDS file defining the rootnames of the process variables. If not specified on the command line, then the namelist commandrootname_listis required in the input file.-echoinput-- echos input file to stdout.-dryrun-- the ``variable'' process variables are left untouched during the execution. The ``measurement'' process variables are still read. The pauses and system calls are still in effect.-summarize-- gives a summary of the experiment before executing it.-verbose[=very]-- prints out information during the execution such as notification of setting and reading process variables. The optionveryprints out the average measurement values.-ezcaTiming=<timeout>,<retries>-- sets EZCA timeout and retry parameters-describeinput-- Printouts the list of namelist commands and fields of the input file.